If you're curious about the link between fungi and cancer, then you need to read this blog!

Introduction
Fungi are understudied but important commensals/opportunistic infections that shape host immunity. Individual tumour types were found to have fungi, which contribute to esophageal and pancreatic cancer carcinogenesis. Their presence, identity, location, and effects in the majority of cancer types are unknown. Fungi and bacteria have symbiotic and antagonistic relationships (Frey-Klett et al., 2011; Peleg et al., 2010; Shiao et al., 2021), which further motivates research into their interactions in tumours. Combining their data results in synergistic colorectal cancer diagnosis.
Here are the key points of this research:
Results
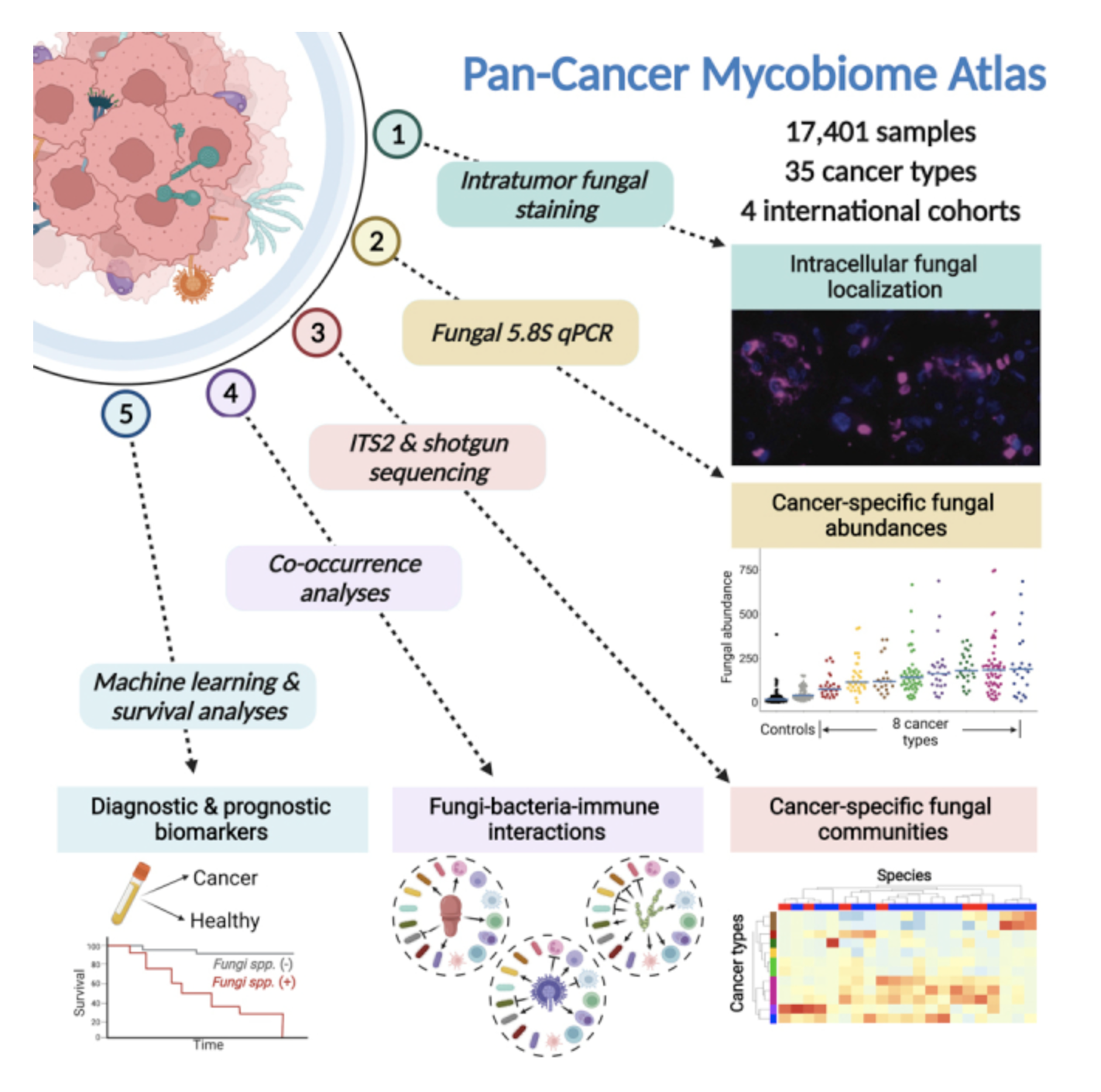
- Many human cancers contain fungus nucleic acids. In two large cohorts of cancer samples the research team previously examined for bacteria, we profiled fungal DNA. Internal transcribed spacer 2 (ITS2) amplicon sequencing was used to check for fungal presence in all samples.
- The second cohort included data from The Cancer Genome Atlas that included whole genome sequencing (WGS) and transcriptome sequencing (RNA-seq). We re-aligned all unmapped DNA and RNA reads to a uniform human reference for quality control.
- The fungal 5.8S ribosomal gene was quantified using quantitative polymerase chain reaction (qPCR) in the WIS cohort. The fungus load in all tumour types was higher than that of negative controls. In colon and lung tumours, the fungus load was significantly higher.
- They found significant, cancer type-specific variations in the percentage of classified fungal, bacterial, and pan-microbial reads in 31 of 32 cancer types. In primary tumours, bacterial read proportions were significantly higher than fungal read proportions. During paired analyses, all cancer types had significantly higher bacterial proportions.
- The scientists calculated per-sample and aggregate fungi genome coverages across all WGS and RNA-seq samples. Saccharomyces cerevisiae, Malassezia restricta, Candida albicans, and Blastomyces gilchristii are among the 31 fungi with R1% aggregate genome coverage.
- Each of the WIS and TCGA cohorts has its own set of advantages and disadvantages. Bacterial genome analyses are affected by differences in sample preparation, sequencing, bioinformatic pipelines, and reference databases. Despite these differences, they discovered 87.2 percent of WIS species and 93.4 percent of fungal genera in matched TCGA cancer types.
- Multiple staining techniques can be used to identify fungi in human tumours.
- By staining melanoma, pancreas, breast, lung, and cervical cancer tissue microarrays, we were able to see fungi in human tumours. They combined four staining techniques with varying degrees of sensitivity and specificity because no staining method can detect all fungi in tissues.
- Overall, 0% to 25% of tumours per cancer type were positive for either b-glucan or Aspergillus staining.
- In WIS and TCGA cohorts, the diversity of the mycobiome varied significantly across cancer types. The bacterial richness of tumours was significantly higher than the fungal richness. Intrauterine fungal and bacterial richness was found to be strongly positive in four of the seven cancer types shared by both cohorts.
Which bacteria or fungi were present?
- Alpha diversity in the WIS intratumoral mycobiome was low, but beta diversity was high between tumour samples. The intratumor mycobiome was dominated by Ascomycota and Basidiomycota phylas across WIS cancer types. Due to abundant Saccharomycetes, the Ascomycota to Basidiomycota ratio (A/B ratio) was greatest in colon cancer (A/B = 8.8) and lowest in melanoma.
Intratumoral mycobiome-bacteriome-immunome interactions
- Fungi and bacteria interact physically and biochemically. Breast cancer, which had the most samples, had the most significant interdomain co-occurrences. Positive fungi-bacteria co-occurrences in breast cancer were found in 96.5 percent of cases.
- The scientists reasoned that intratumoral fungal-bacterial-immune clusters exist because fungi and bacteria produce distinct host immune responses. In the TCGA, we compared WIS-overlapping fungal and bacterial genera with TCGA immune cell compositions.
- Unsupervised analyses revealed three distinct fungi-bacteria immune clusters driven by fungal co-occurrences. Raw counts were then aggregated within each domain (e.g., bacteria) and mycotype to create log-ratio comparisons.
- The research team then looked into whether mycotypes were linked to immune responses previously seen in TCGA patients. Immune cells that co-occur with F1, F2, or F3-clustered fungi have significantly different log ratios. Different intratumoral mycobiomes may elicit different host responses. Cancer-type-specific tissue and blood mycobiomes are supported by statistical and machine-learning analyses.
Clinical utility of cancer mycobiomes
- The cancer mycobiome's diagnostic and prognostic capabilities were then investigated. The research team first looked at fungal associations with disease phenotypes using the WIS cohort.
- Cladosporium sphaerospermum and the Cladosporidium genus, previously reported in breast cancer, were found to be highly concentrated in tumours of women aged R50 years old. Cladosporium was also found to be abundant in human epidermal tissue. They discovered increased intratumoral fungal abundance and enrichment of Aspergillus in lung cancer.
- TCGA's blood-derived, stage-invariant, cancer-type-specific fungal compositions suggest their utility as minimally invasive diagnostics. These findings were validated in two independent, published cohorts (Hopkins, UCSD) with 330 healthy and 376 cancer-bearing individuals.
- Human-read removal, microbial classification, and fungal decontamination were all performed in the Hopkins cohort (n = 537; 8 cancer types). They used a published machine learning framework and hyperparameters to assess pan-cancer versus healthy diagnostic performance of raw microbial abundances.
- ML analyses were focused on Hopkins' 45 stage I, treatment-naive samples from eight cancer types. Decontaminated fungi species performed well.
- All 209 Hopkins cohort decontaminated fungi performed at least as well as the top 20 ranked, decontaminated fungal species (9.6% of total). These 20 fungi also distinguished well among batch-corrected, pan-cancer TCGA blood samples (AUROC 95 percent CI: [87.76, 89.79]%; Data S6.2F) When compared to negative controls, performances with this signature or all decontaminated moulds in the UCSD cohort revealed expected results.
Discussion
- In four separate cohorts across 35 cancer types, the scientists investigated the mycobiomes of 17,401 tissue and blood samples. Although fungi were found in all cancer types examined, not all tumors were found to be positive for fungal signal.
- Malassezia globosa promotes pancreatic oncogenesis.
- Patient survival was stratified by fungus "mycotypes" with different immune responses. Fungi are rare but immunologically potent, according to data.
- Across several cancer types, the team discovered strong positive correlations between fungal and bacterial diversity, abundance, and co-occurrences. This suggests that tumour microenvironments (TMEs) may be non-competitive environments for multi-domain microbial colonization.
Plasma mycobiomes are analysed for the first time in treatment-naive, early-stage cancers. Although the sources of cell-free plasma-derived fungal or bacterial DNA are unknown, they are likely.
Implications
- Intramuscular fungal presence and tumour-specific localisation patterns were discovered using four different staining techniques.
- Staining techniques can only detect a small portion of the fungi kingdom.
- This is the first pan-cancer mycobiome atlas.
- It adds a new layer of information to cancer diagnostics and treatment.
Reference:
Narunsky-Haziza L, Sepich-Poore GD, Livyatan I, Asraf O, Martino C, Nejman D, Gavert N, Stajich JE, Amit G, González A, Wandro S, Perry G, Ariel R, Meltser A, Shaffer JP, Zhu Q, Balint-Lahat N, Barshack I, Dadiani M, Gal-Yam EN, Patel SP, Bashan A, Swafford AD, Pilpel Y, Knight R, Straussman R. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 2022 Sep 29;185(20):3789-3806.e17. doi: 10.1016/j.cell.2022.09.005. PMID: 36179670; PMCID: PMC9567272. https://pubmed.ncbi.nlm.nih.gov/36179670/
Video coming soon...